You are viewing the RapidMiner Radoop documentation for version 9.2 - Check here for latest version
Advanced Radoop Processes
To understand advanced Radoop process design, you should be familiar with the RapidMiner data flow, Radoop basics, and how to import data. The following section introduces the predictive analytics capabilities of RapidMiner Radoop and illustrates how smoothly it integrates into the RapidMiner client interface by using the same I/O objects, metadata, and process design concepts. However, because Radoop implements operations in Hadoop, you can deploy your process (or a selected part of it) on the cluster to manage the enormous data volumes using scalable distributed algorithms.
Model objects
Data mining methods, primarily supervised learning algorithms, create prediction models. (Other algorithms generate other types of models, but this section focuses mainly on prediction and clustering models.) These models describe the information explored from a training data set. A RapidMiner model is an I/O object — a Prediction Model, Cluster Model, or Preprocessing Model — just like an ExampleSet object. Visualization of the model displays the explored information to the user.
Both Radoop and core RapidMiner operators train and apply the same types of model objects. More precisely, Radoop implements the scoring and learning of some popular RapidMiner models over the distributed infrastructure; all core prediction models can be applied now on the cluster. This is true not only for prediction models, but also for the preprocessing models that a preprocessing operator, for example, Add Noise, generates (Noise Model).
Another type of I/O object related to modeling is the Performance Vector object. Predictions, in the form of models, are evaluated by comparing the expected target attribute values to the result of the scoring. With that result, performance criteria create measures to describe the validity of the prediction (model) from the viewpoint of the data mining process goals. In other words, Radoop performs — on the cluster — scoring and model application as well as performance criteria calculation.
Data mining scenarios
This section reviews some possible memory/cluster scenarios for your predictive modeling processes.
Unsupervised learning in Hadoop. In this scenario, scalable distributed algorithms create clustering of data in Hadoop. Some clustering algorithms build clustering models that you can apply on other similar data sets, either on Hadoop or in memory. The model is delivered on the cluster model (clu) output port of clustering operators.
Supervised learning in memory, scoring on the cluster. Core RapidMiner modeling (classification and regression) operators build predictive models in memory that you can then apply to your data in the cluster. Radoop supports applying all core prediction models. By using the SparkRM or Single Process Pushdown operators the learning can be performed in the memory of one or multiple cluster nodes in parallel using any core RapidMiner modeling operator.
Supervised learning and scoring with native algorithms on the cluster. In this scenario, learning and scoring are both implemented on the distributed platform. Radoop supports the following algorithms: Naive Bayes, Linear Regression, Logistic Regression, Support Vector Machine, Decision Tree, and Random Forest. It also supports iterative Naive Bayes model building, which means that a model (built on the cluster or in memory) can be updated by new data (on the cluster or in the memory).
Clustering models
Radoop clustering operators are built on the distributed machine learning algorithms of the Apache Mahout project. The input of a clustering operator is a HadoopExampleSet object. The algorithm creates the specified number of segments in the data. It adds a new attribute to the data set, named clusterid, that has the cluster role. The attribute contains the cluster identifier cluster_1, cluster_2, etc.
Clustering operators may also indicate that a record is so distant from the rest of the records that it should be considered an outlier. In this case, the cluster attribute contains the value outlier.
K-Means and Fuzzy K-Means algorithms also build a centroid clustering model. You can use the centroid model to cluster similarly structured data sets that reside on your cluster or in memory.
Classification and regression models
You can connect prediction and cluster models trained by core RapidMiner operators in the client machine's memory or in a cluster node's memory (using the SparkRM or Single Process Pushdown operators) to the input port of a Radoop Nest and use them inside its subprocess. Radoop's Apply Model operator takes just such a model and a test data set as inputs. The operator applies the prediction model on the HadoopExampleSet input and delivers a HadoopExampleSet with new prediction and confidence columns. In a cluster model, Radoop generates a cluster column.
Radoop supports scoring on the cluster for all RapidMiner prediction and cluster models. These models produce the same prediction inside and outside the nest on the same data sets, but scoring inside the nest is not limited to the operative memory size; it is scalable over the distributed platform.
You can also train a prediction model on the distributed platform. Radoop supports Naive Bayes, Linear Regression, Logistic Regression, Support Vector Machine, Decision Tree and the Random Forest learning algorithms. These algorithms have the following characteristics:
| Algorithm | Attribute type | Class type | Notes |
|---|---|---|---|
| Naive Bayes | numerical and nominal | polynominal | Contains a unique, linearly scalable implementation for Naive Bayes. Ideal for iterative learning. |
| Linear Regression | numerical | numerical and binominal | Integrates the Spark MLlib implementations. |
| Logistic Regression | numerical | binominal | Integrates the Spark MLlib implementations. |
| Support Vector Machine | numerical | binominal | Integrates the Spark MLlib implementations. |
| Decision Tree | numerical and nominal | binominal and polynominal | The Decision Tree (MLlib binominal) integrates the Spark MLlib implementation and can handle binominal label, the Decision Tree operator handles polynominal label and integrates spark.ml's algorithm. |
| Random Forest | numerical and nominal | polynominal | Integrates spark.ml's Random Forest algorithm. |
You can apply models trained with the above algorithms in the same way, using the core Apply Model operator (in-memory) and the Radoop Apply Model operator (in-Hadoop).
Iterative learning
Iterative learning is a special type of learning for classification or regression tasks. With each iteration, Radoop updates the model with a new training data set. Since the much faster update with the new records results in the same model, you can rebuild the model with just the new records as if you had built it with the whole new data set. This is a very common scenario for big data analytics (for example, new log records may update a prediction model periodically).
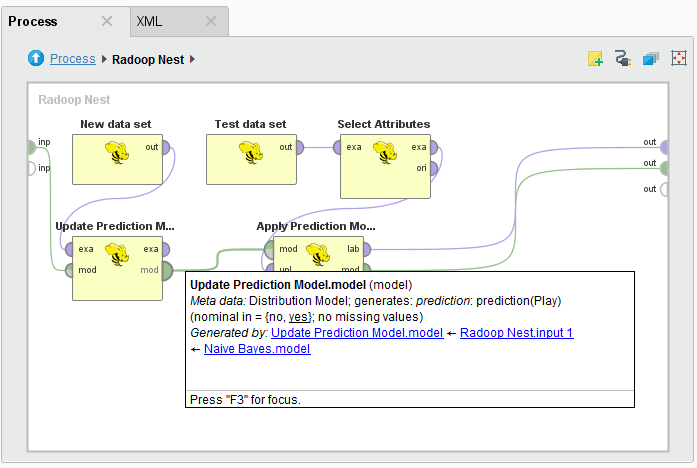
Radoop implements iterative learning with the Update Model operator. This operator takes a model as input (a Distribution Model that was built with Naive Bayes) and updates it by training on the HadoopExampleSet on its other input port. The input model may have been trained either by a Radoop learner operator (an operator that creates a model based on an ExampleSet, such as Naive Bayes) or by a core RapidMiner learner. The new data set must have exactly the same schema as the original training data set. That is, the name and attribute type must match, but ordering is irrelevant. The learning algorithm of the Update Model operator may (or may not) have parameters, for example optimization parameters. Therefore, the Update Model operator has a special "generic" parameter that allows you to easily specify model type specific parameters. (For example, if Update Model has a DistributionModel on the input, you can set the nominal group size training parameter.)
The following illustrates model update and apply in a single subprocess:
Ensemble modeling
Radoop contains a meta operator called Combine Models. This operator can be used both inside and outside of the Radoop Nest. It simply creates a Bagging Model — sometimes called a voting model — out of the trained models on its input. (Note that all input models have to be trained on data with the same schema.) The input can be a collection of models or an arbitrary number of models on the input ports. The ensemble model on the output port will be a voting model. If this model is used for scoring, it applies all inner models, then takes the average of confidence values (if it's a classification model) to decide the predicted class. In the case of a regression model (having a numerical label), it calculates the average of the inner models' prediction.
You may want to use the Combine Models operator together with a training loop if you have too much training data on the cluster and, for some reason, the distributed algorithms provided by Radoop do not fit your use case. You can fetch random samples from your data to memory and train different models on these samples. After combining these models into a single Bagging Model, you can then apply the model directly to your data on the cluster. In this way, you not only use RapidMiner's wide range of learning algorithms, but you build a very precise and robust ensemble model in a short time using only samples of data.
You can also use only a subset of the regular attributes in each training iteration to build a more precise voting model. The Select random attributes operator is ideal for this task. For the training algorithm, RapidMiner Radoop's decision tree learning algorithm may be a good choice. This way, you can implement, for example, a Random Forest learning algorithm, controlling the sample size, the number of iterations, and the randomization method based on the resources available (memory, bandwidth, amount of data). You can use any unstable learning algorithm instead of Decisision Tree to benefit from the Combine Models operator, as Radoop support building the Random Forest model in a distributed way with a single operator.
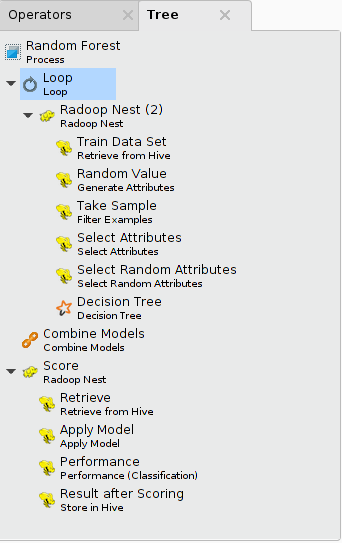
The following illustrates a process that implements a Random Forest training algorithm on data samples (a loop with 100 iterations). It uses the Tree panel of the process to show all operators. You can open the Tree panel for a process by selecting the View > Show Panel > Tree menu item.
In-memory subprocesses
RapidMiner Radoop offers multiple ways to take advantage of RapidMiner's in-memory operators. The In-Memory Subprocess (Sample) and In-Memory Subprocess (Full) operators implement the concept described in the previous section in a more general way. They both operate on subsets of rows. These operators are very useful for training ensemble models, but In-Memory Subprocess (Full) is also capable of doing other preprocessing tasks. Unlike the In-Memory Subprocess, the SparkRM and Single Process Pushdown operators are capable of executing the subprocess on a cluster node using the node's available memory. This way you can use the Hadoop cluster's resources to perform any in-memory computation task.
Using the cluster's memory
The SparkRM and Single Process Pushdown operators can be used to execute in-memory subprocesses on one or more cluster nodes. They are described on details on the Process Pushdown subpage.
Using the client machine's memory
Since large datasets reside on the cluster, and cluster nodes are expected to have more resources than a client machine (that runs Studio or Server in this case), SparkRM and Single Process Pushdown should be preferred over the two (in a sense, legacy) operators described below.
In-Memory Subprocess (Sample) is a meta operator inside the Radoop Nest that runs its subprocess on a data sample in operative memory. The Radoop process fetches a sample from the HadoopExampleSet on the In-Memory Subprocess (Sample) input port to memory, and the subprocess — consisting of any core RapidMiner operators — is applied on this data. The data is then written back to the HDFS for further processing by the Radoop process. You can connect other I/O objects, for example a model trained on this sample data, to the output port for later use on the cluster. Please note that the SparkRM or Single Process Pushdown operators are preferred over the In-Memory Subprocess (Sample) as they perform the same operation but use the cluster's resources instead of the client machine's.
In-Memory Subprocess (Full) differs from the sample version in the following way: it processes all the data on its input port in small partitions that fit into operative memory. You must either define the number of these partitions that the input data will be split into or the size of the partition. The meta operator performs a loop on these partitions. In each iteration it reads the data in the current partition to memory, runs the subprocess on it, and then writes the data back to the HDFS (appends it to the output table).
There are basically two different scenarios for using the In-Memory Subprocess (Full) meta operator:
when you want to do special data preprocessing that you can not implement with Radoop operators, but can do with core RapidMiner operators (or operators from another extension). In this case, with the meta operator you can stream your data through the client's operative memory while preprocessing it. As the operative memory and/or network bandwidth may limit this, larger data sets are best served using RapidMiner Server. Your server may have enough memory and a good connection to process larger input data (larger partitions mean fewer iterations).
when you build object(s) using splits and you do not need to write the data back to the cluster. For example, similar to the Random Forest example in the previous section, you can train as many Decision Tree models to as many partitions as you choose (that is, "number of decision trees on the output" = "number of partitions"). You can then combine these models to a voting model with the Combine Models operator. To do this, connect the model built inside the In-Memory Subprocess (Full) meta operator to the output, then connect the <Collection of Decision Tree> output of the meta operator to the Combine Models input. This way, you train a model on data that is much larger than the operative memory size, but can still expect the voting model to have quite good precision.
Just like In-Memory subprocess (Sample), In-Memory Subprocess (Full) can be replaced by the Single Process Pushdown. If your data is bigger than the memory of the largest node in your cluster, you can use the Generate Attributes operator to create a random attribute and Filter Examples in a Loop to create the partitions for the Pushdown operator.
Evaluation
The purpose of performance evaluator operators is to provide simple measures for evaluating the current prediction and the prediction model itself. Analysts can define the target measures that they want to optimize for and the models can be compared using these performance criteria.
Radoop implements measures for evaluating binominal and polynominal classification and regression. These form a subset of the set of criteria that core RapidMiner evaluation operators implement. The following list of performance criteria describes these measures.
| Type | Criterion name | Criterion description |
|---|---|---|
| binominal and polynominal | accuracy | Relative number of correctly classified examples |
| binominal and polynominal | classification_error | Relative number of misclassified examples |
| binominal and polynominal | kappa | Kappa statistics for the classification |
| binominal | precision | Relative number of correctly as positive classified examples among all examples classified as positive |
| binominal | recall | Relative number of correctly as positive classified examples among all positive examples |
| binominal | lift | The lift of the positive class |
| binominal | fallout | Relative number of incorrectly as positive classified examples among all negative examples |
| binominal | f_measure | Combination of precision and recall: f=2pr/(p+r) |
| binominal | false_positive | Absolute number of incorrectly as positive classified examples |
| binominal | false_negative | Absolute number of incorrectly as negative classified examples |
| binominal | true_positive | Absolute number of correctly as positive classified examples |
| binominal | true_negative | Absolute number of correctly as negative classified examples |
| binominal | sensitivity | Relative number of correctly as positive classified examples among all positive examples (same as recall) |
| binominal | specificity | Relative number of correctly as negative classified examples among all negative examples |
| binominal | youden | The sum of sensitivity and specificity minus 1 |
| binominal | positive_predictive_value | Relative number of correctly as positive classified examples among all examples classified as positive (same as precision) |
| binominal | negative_predictive_value | Relative number of correctly as negative classified examples among all examples classified as negative |
| binominal | psep | The sum of the positive predictive value and the negative predictive value minus 1 |
| polynominal and regression | absolute_error | Average absolute deviation of the prediction from the actual value |
| polynominal and regression | relative_error | Average relative error (average of absolute deviation of the prediction from the actual value divided by actual value) |
| polynominal and regression | relative_error_lenient | Average lenient relative error (average of absolute deviation of the prediction from the actual value divided by maximum of the actual value and the prediction) |
| polynominal and regression | relative_error_strict | Average strict relative error (average of absolute deviation of the prediction from the actual value divided by minimum of the actual value and the prediction) |
| polynominal and regression | root_mean_squared_error | Averaged root-mean-squared error |
| polynominal and regression | squared_error | Averaged squared error |